Abstract:
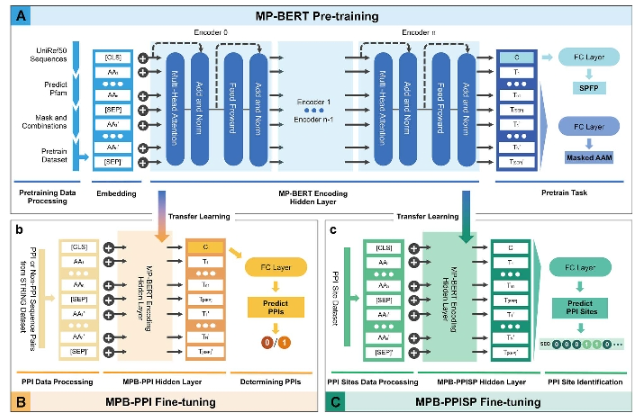
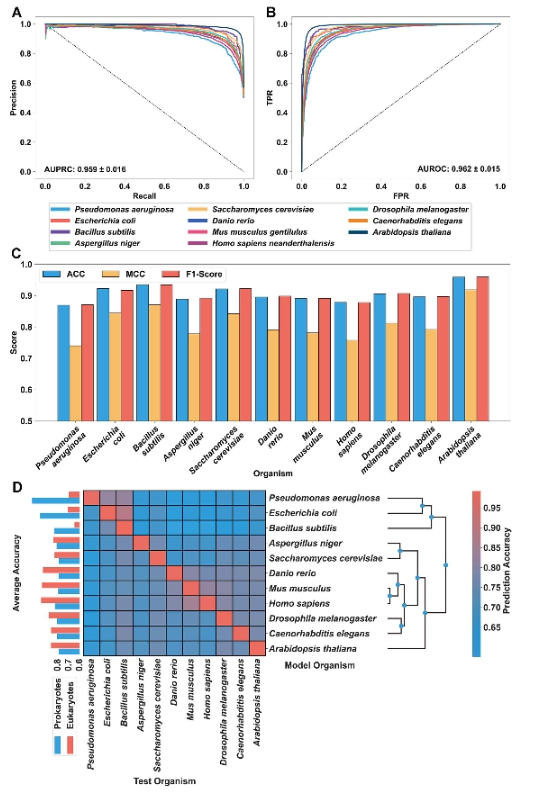
The advanced language models have enabled us to recognize protein-protein interactions (PPIs)and interaction sites using proteinsequences or structures.Here, we trained the Mindspore ProteinBaKT MP-BEK model, a Bidirectional Encoder Representation fromTransformers.jusing protein pairs as inputs, making it suitable for identifying PPis and their repective interaction sites. The pretrainedmod 查大T) was fine-tuned as MPB-PPI(MP-BERT on PP and demonstrated its superiority over the state-of-the-art models ondiverse dencimark datasets for predicting PPls.Moreover, the model's capability to recognize PPls among various organisms wasevaluated on multiple organisms. An amalpamated orpanism model was designed, exhibiting a high level of generalization acrosshe majority of organisms and attaining an accuracy of 92.65%. The model was also customized to predict interaction site propensityby fine-tuning it with PPI site data as MPB-PPISP Our method facilitates the prediction of both PPIs and their interaction sites, therebyillustrating the potency of transfer learning in dealing with the protein pair task.
Key Words:
protein-protein interaction; transformer; PPI site; transfer learning; BERT